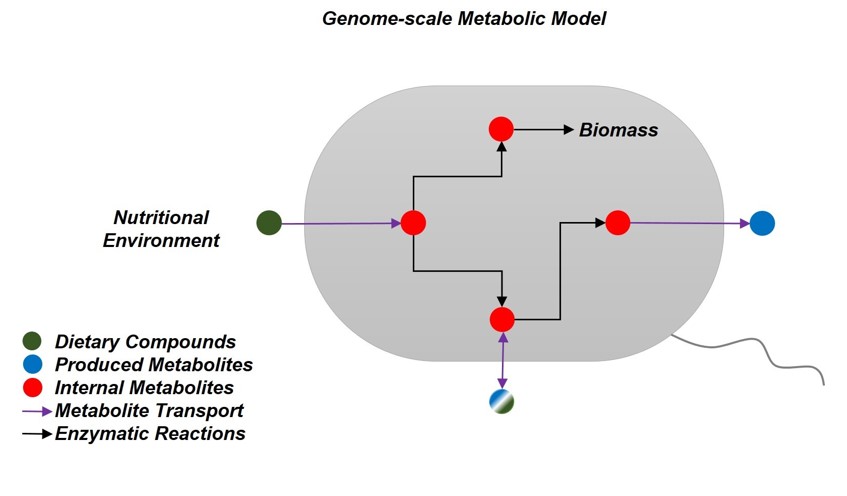
Genome-scale metabolic modeling: How to model the nutritional environment
Genome-scale metabolic modeling is a computational technique that enables the simulation of the metabolism of living cells. The models are based on metabolic networks which describe the biochemical capabilities of an organism to use environmental resources for cellular growth. Thus, genome-scale metabolic network modelling has been established as a key tool in a wide number of fields including medicine, molecular biology, and ecology. A crucial step towards using these networks in simulations of cell metabolism is to model the nutritional environment of the cells. However, this step is usually complicated and can become overwhelming, because the availability of nutrients often cannot be accurately estimated. The main reasons for this issue are the frequent use of complex constituents for bacterial growth media whose composition varies (e.g. of yeast), or the lack of knowledge of the specific nutritional environment of a bacterial species (e.g. in the gastrointestinal tract of rodents).
To overcome these problems, the research groups of Prof. Dr. Christoph Kaleta -a CRC1182 member- and Prof. Dr. Silvio Waschina have recently published an article that provides guidelines on defining the nutritional environment for metabolic models. This publication serves as a reference for both beginners and experienced researchers in the field, as it describes the required procedures step-by-step and in detail. Furthermore, it provides the readers with a realistic and common example; that is the modeling of the Lysogeny Broth complex medium for an in-silico simulation of the metabolism of a Escherichia coli metabolic model. Thus, this work represents an important step forward in improving the accuracy of metabolic modelling by providing a guideline on how to more specifically determine the nutritional environment of the modeled organisms.
Article: Marinos G, Kaleta C, Waschina S (2020) Defining the nutritional input for genome-scale metabolic models: A roadmap. PLOS ONE 15(8): e0236890. https://doi.org/10.1371/journal.pone.0236890